Computational Biology & Bioinformatics Laboratory

Computational Biology and Bioinformatics is a rapidly developing multidisciplinary field. There has been a great increase in the amount of biomedical data over the past decade. Along with the expanding application of large-scale genomic sequencing, other modalities such as mobile health (mHealth) data and imaging have added to the rise. At the same time, computing power and storage capacity have continued to increase, allowing us to now mine and model biological data with unprecedented ability. Our research activities include computational modeling of biological processes, computational management of large-scale data sets, database development and data-mining, algorithm development and high-performance computing, as well as statistical and mathematical analyses.
Faculty Members associated with the lab

Sushmita Paul
Assistant Professor
Pankaj Yadav
Assistant ProfessorGroups under this theme
1. Computational Biology Group |
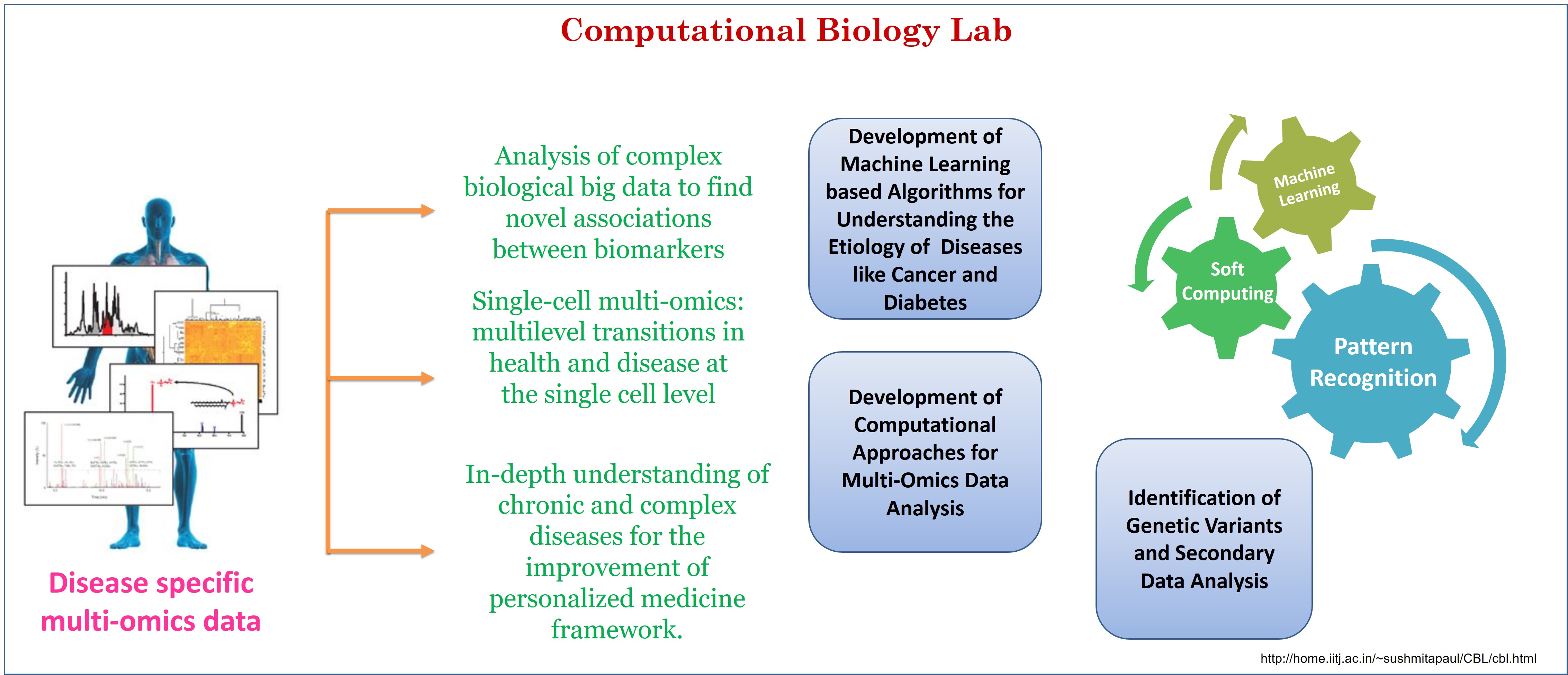
| Computational Biology Lab (CBL) is established to carry out fundamental and advanced research in the fields of computational biology and bioinformatics. Dr. Sushmita Paul's research group carries out research in multi-omics data analysis, development of pattern recognition algorithms for analysing high dimensional biological data, analysis of genome variation, development and application of bioinformatics tools. The group is actively involved in development of algorithms for identification of miRNA-mRNA modules in various diseases by using multi-omics data. Another important challenge related to multi-omics data analysis is classification of cancer subtype. In this regard, the group has developed an algorithm for effectively classifying the cancer samples into their respective sub-types. The group is also involved in functional annotation of genomic variants in Indian population, sub-grouping of indian population based on genetic variants. The group is also focusing on development of an AI based framework to predict patient outcome to treatment based on patient biopsy derived tumour spheroid. The group also developed several algorithms/frameworks to identify Type II diabetes genes by judiciously integrating gene expression data and protein-protein interaction network data. The CB Lab also conducted an international workshop on Recent Advances in Biomedical Data Analysis at Olsztyn, Poland, 2017 (http://ijcrs2017.uwm.edu.pl/?page_id=190). In 2019, the lab organized a national level workshop on Computational Biology and Bioinformatics at IIT Jodhpur (http://home.iitj.ac.in/~sushmitapaul/Workshop2019/). |
|
2. Life Science Informatics & Statistics Group
|
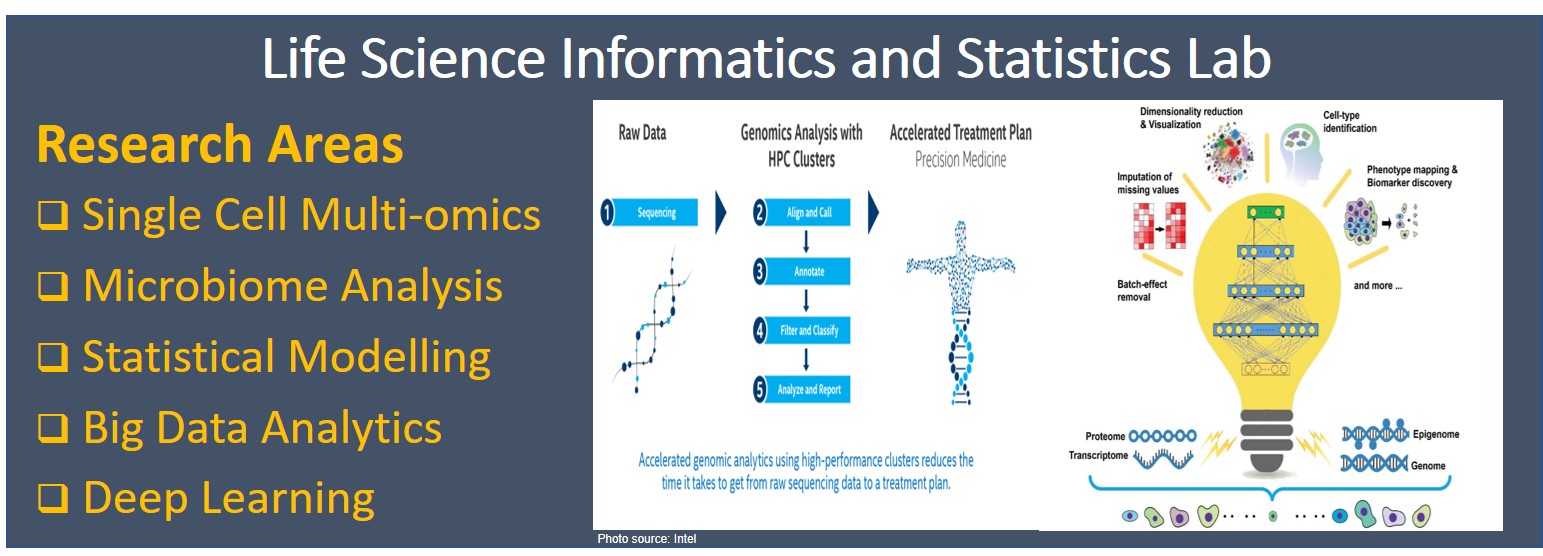
Recent advancements in technologies have generated huge amounts of biological and clinical data for researchers. This wealth of data poses challenges that have never before been confronted. At the heart of these is understanding how massive biological data sets are best analyzed to discover new knowledge about the function of living systems in health and disease, and how this knowledge can be harnessed to provide improved, more affordable health care. To this end, sophisticated tools are needed to manage and analyse such a large volume of the data sets. This research group is dedicated to develop advanced statistical and computational methods for drawing statistically valid inference from biological and clinical data. We study inter-individual differences by large-scale statistical modelling and integrating multiple layers of OMICS data. |
Projects that the lab is currently catering to |
1. Analysis of miRNA and mRNA Expression Data to Understand Etiology of Cervical Cancer (Dr. Sushmita Paul) |
Cancer is the second most common cause of death in India. More women in India die from cervical cancer than in any other country. In order to address the challenges of cancer few projects have been initiated internationally like The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC). These projects use advanced genomic technologies to generate genomic data that are used to draw statistically and biologically significant conclusions. Although many important discoveries have been made by TCGA's research network, opportunities still exist to implement novel methods, thereby elucidating important diagnostic markers and new biological pathways. The main objective of the project is to develop some computational approaches to understand the role of multimodal omics data, that is, miRNAs and mRNAs responsible for genesis and progression of cervical cancer. In this regard, an unsupervised clustering algorithm will be developed that may help to characterize molecular subtypes of cancer for an unlabeled data. A supervised clustering method will be developed for identification of clusters of correlated biomarkers that are strongly associated with sample categories. Simultaneous feature selection and clustering algorithms will be developed for grouping different types of biomarkers that exhibit similar behavior across a subset of samples or clinical outcomes. Finally, the biological significance of the obtained results will be studied using gene ontology, network analysis, and wet lab experiments, as well as the biological information available in the literature. |
| 2. Integrative Approach for Identification of Disease Genes of Type II Diabetes (Dr. Sushmita Paul) |
| Currently, India is facing diabetes as a serious epidemic condition. About 62 million diabetic individuals are already diagnosed with the disease. In a survey it was observed that India (31.7 million) has the maximum number of cases with diabetes mellitus followed by China (20.8 million) and USA (17.7 million) in second and third position, respectively. The number of diabetic patients will be doubled by 2030 worldwide and India will have the maximum number of cases. If immediate action is not taken for understanding aetiology of the disease then India may face potential burden. Many genetic/epigenetic factors are responsible for the prevalence of diabetes and identification of those factors is important to understand the biology of the disease. Interaction between identified genetic factors is also needed to be studied in order to understand incidence of the disease. A potential disease gene of diabetes may be used as a target for developing drugs against it. Also, the disease genes may be used for development of diagnostic tools that may help to understand the chances of occurance of the disease. However, identification of potential disease genes experimentally is very costly as well as requires a lot of time. In order to overcome this issue statistical and computational methods can be used to identify a list of potential disease genes. Predicted disease genes will not only reduce cost of experiment but also time requirement will also get reduced. Network analysis of the predicted genes will help to understand the mechanism of the disease. |
| 3. GenomeIndia: Cataloguing the Genetic Variation in Indians (Dr. Sushmita Paul) |
| Genetic variations in individuals are often population specific and could predispose them to diseases and determine their response or adverse effects to certain drugs. This has led several nations to implement population-specific sequencing projects. Despite being a large population with diverse ethnic groups, India lacks a comprehensive catalogue of genetic variations. In India, we do not have a reference catalog of genetic variations, which makes identifying the causal variations for monogenic disorders difficult and inaccurate. Currently, India also does not have a country specific genome wide chip that will make large scale comprehensive genetic studies affordable. This calls for a national level effort in India to catalogue the genetic variations of its diverse population by whole genome sequencing of a large number of individuals from diverse geographical regions and ethnicities spanning the country. The outcome of this study would also be useful to other South Asian countries, as Indian genetics is relevant to the neighboring South Asian countries. This will be an immense gain of knowledge and information for the world-wide human genetics research community as large scale comprehensive genetic studies have conventionally centered around European populations, in general, and only recent inroads have happened for African and Hispanic populations. |
| 4. Development of an AI platform for human health (Dr. Sushmita Paul, Co-PI) |
| To develop an AI based framework to predict patient outcome to treatment based on patient biopsy derived tumour spheroid. To develop an automated chest radiograph based diagnostic tools for detection and differentiation of silicosis and tuberculosis. |
| 5. Investigation of genome-wide gene-environment interactions in relation to complex diseases following case-only approach (Dr. Pankaj Yadav) |
| Non-communicable diseases (NCDs), particularly cancer, chronic respiratory diseases and diabetes, are the leading causes of mortality globally. Of the 56 million global deaths in the year 2012, 38 million (about 68%) were due to NCDs and this number is projected to increase to 52 million by 2030. In India, deaths from NCDs are projected to almost double from about 4.5 million in 1998 to 8 million by 2020. This project aims to implement the case-only approach to the available data to detect underlying gene-environment interaction at the genome-wide level in relation to different NCDs. |
Key Instrumentation Available |
| Servers |
| Workstations |
| Desktops |
Software Developed |
| RFCM3 (http://home.iitj.ac.in/~sushmitapaul/CBL/softwares.html) |